Hepatitis: From dysregulated
lipid metabolism to lasting liver failure
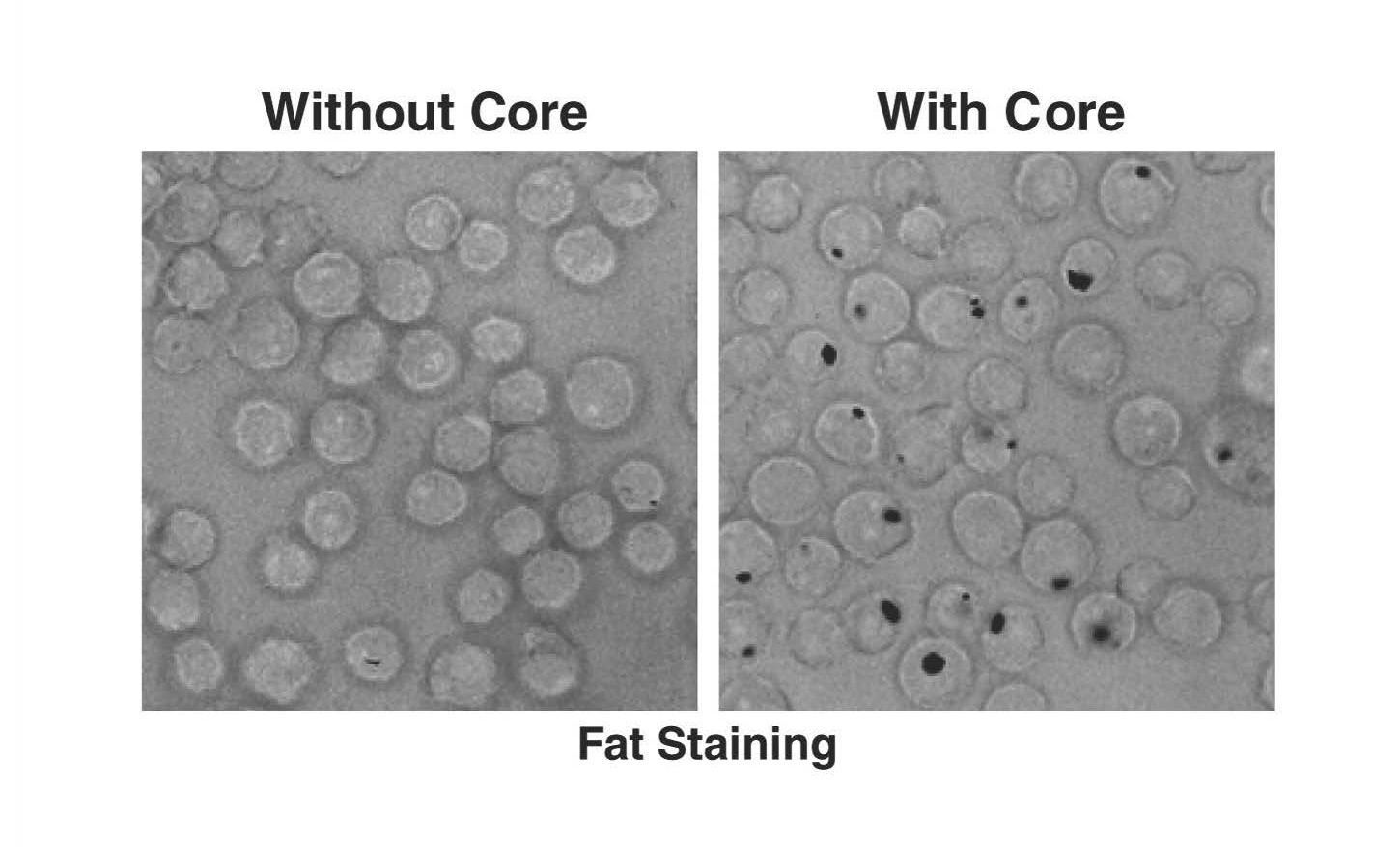
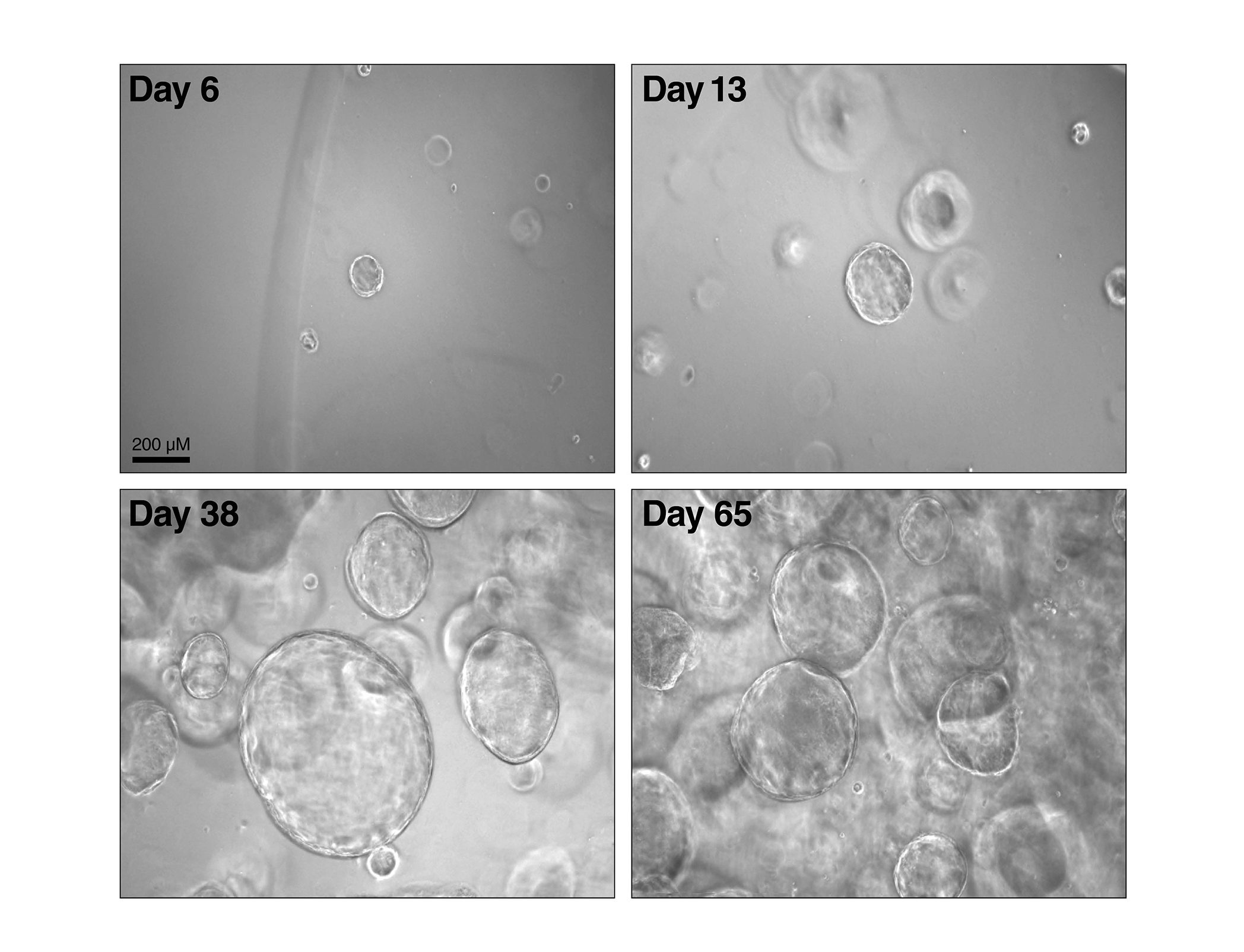
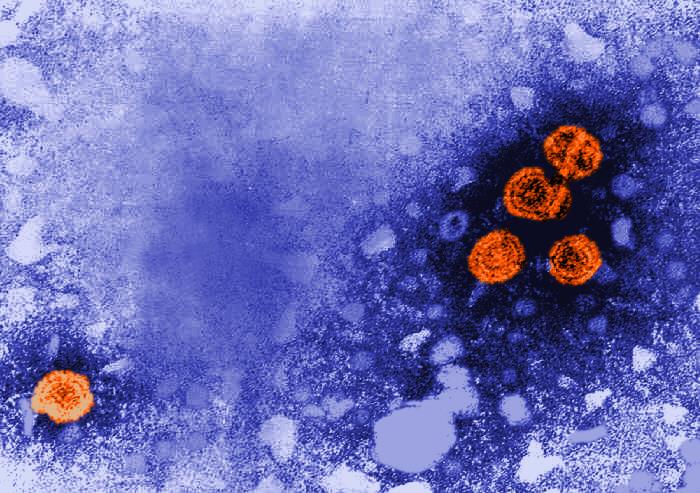
According to the World Health Organization, 1.3 million people die each year of hepatitis. In the United States, liver damage due to chronic infection with hepatitis B or C viruses is the leading cause of liver transplants. Our lab studies the three main hepatitis viruses (HAV, HBV and HCV), with a special focus on fat metabolism in liver cells and the use of 3D liver models to study spreading infection.